GitHub - petosa/simple-alpha-zero: Clean, tested, & modular
Por um escritor misterioso
Last updated 28 dezembro 2024
Clean, tested, & modular AlphaZero implementation with multiplayer support. - GitHub - petosa/simple-alpha-zero: Clean, tested, & modular AlphaZero implementation with multiplayer support.

Comprehensive structure and functional adaptations of the yeast nuclear pore complex - ScienceDirect

Comprehensive Structure and Functional Adaptations of the Yeast Nuclear Pore Complex

A practical guide for analysis of histone post-translational modifications by mass spectrometry: Best practices and pitfalls - ScienceDirect

Comprehensive Structure and Functional Adaptations of the Yeast Nuclear Pore Complex
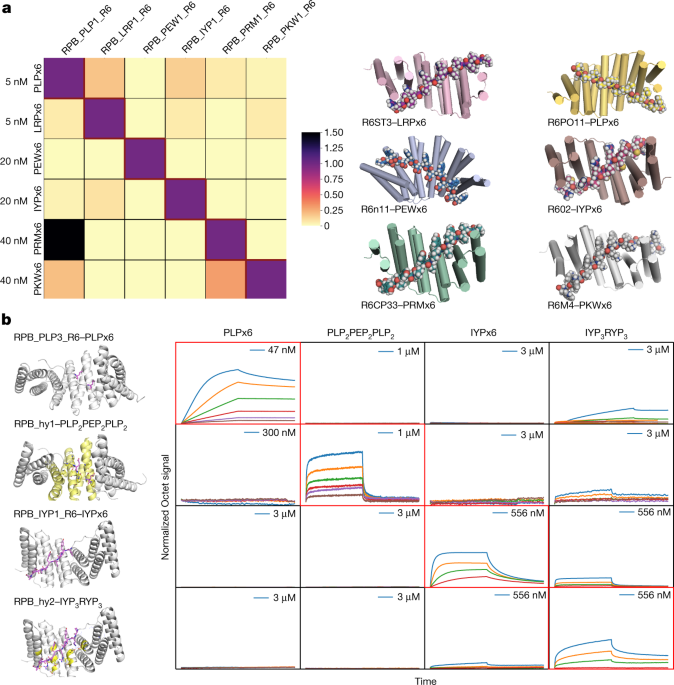
De novo design of modular peptide-binding proteins by superhelical matching
GitHub - alexandersoen/alpha-tree-fair-wrappers: Python implementation of Fair Wrappers for Black-box Predictions

Genome-wide survey of D/E repeats in human proteins uncovers their instability and aids in identifying their role in the chromatin regulator ATAD2 - ScienceDirect

Comprehensive Structure and Functional Adaptations of the Yeast Nuclear Pore Complex

Proteome‐scale mapping of binding sites in the unstructured regions of the human proteome
Recomendado para você
-
 Leela Chess Zero: AlphaZero for the PC28 dezembro 2024
Leela Chess Zero: AlphaZero for the PC28 dezembro 2024 -
 Simple Alpha Zero28 dezembro 2024
Simple Alpha Zero28 dezembro 2024 -
GitHub - Yangyangii/AlphaZero-connect6: DeepMind AlphaZero for28 dezembro 2024
-
alphazero · GitHub Topics · GitHub28 dezembro 2024
-
GitHub - suragnair/alpha-zero-general: A clean implementation28 dezembro 2024
-
 Alpha Zero will be coming back! Who will be the boss , SF 1028 dezembro 2024
Alpha Zero will be coming back! Who will be the boss , SF 1028 dezembro 2024 -
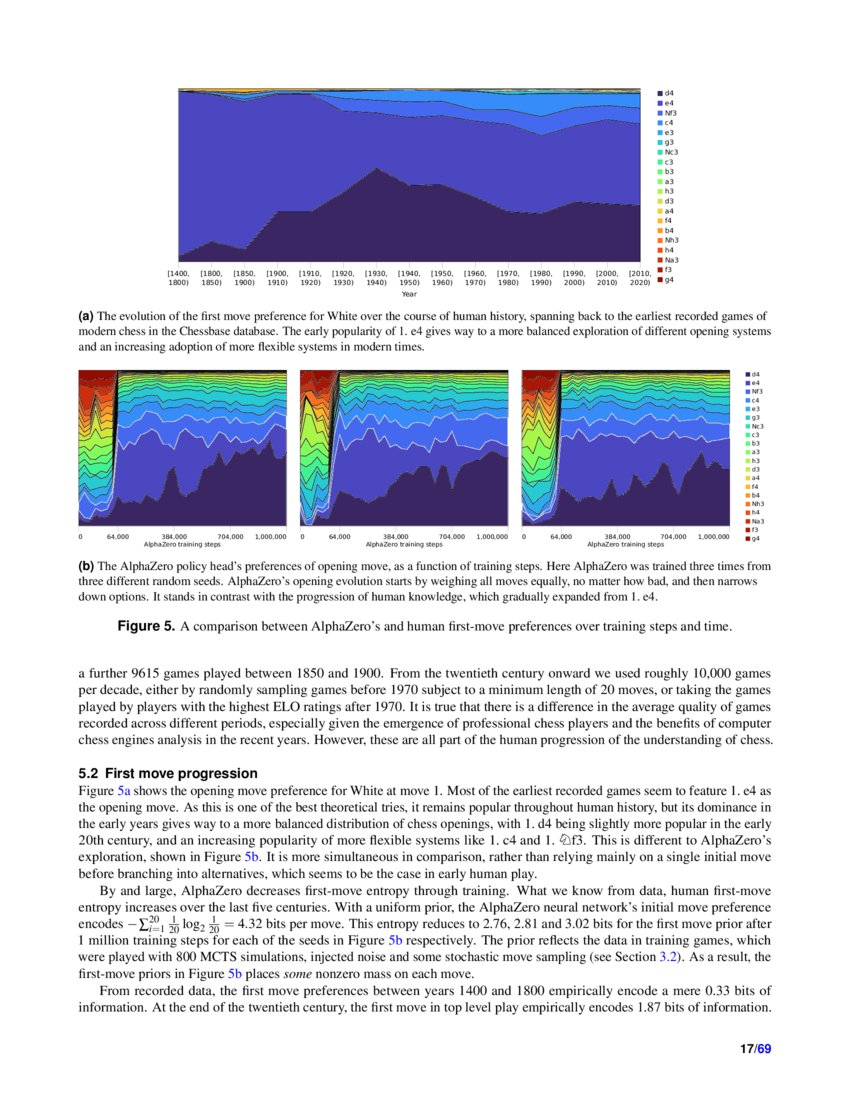 Acquisition of Chess Knowledge in AlphaZero28 dezembro 2024
Acquisition of Chess Knowledge in AlphaZero28 dezembro 2024 -
 Recreating DeepMind's AlphaZero - AI Plays Connect 4 - Part 228 dezembro 2024
Recreating DeepMind's AlphaZero - AI Plays Connect 4 - Part 228 dezembro 2024 -
 Mastering TicTacToe with AlphaZero28 dezembro 2024
Mastering TicTacToe with AlphaZero28 dezembro 2024 -
 What is Q*? And when we will hear more? - Community - OpenAI28 dezembro 2024
What is Q*? And when we will hear more? - Community - OpenAI28 dezembro 2024
você pode gostar
-
 Chad tier list Brawl Stars Amino28 dezembro 2024
Chad tier list Brawl Stars Amino28 dezembro 2024 -
 Boneca Barbie Grávida E Família Bebê Carrinho Genérico28 dezembro 2024
Boneca Barbie Grávida E Família Bebê Carrinho Genérico28 dezembro 2024 -
 This might be the most understandable rage quit in Street Fighter 5 history28 dezembro 2024
This might be the most understandable rage quit in Street Fighter 5 history28 dezembro 2024 -
 SÃO PAULO, SP - 05.03.2022: SÃO PAULO FC X CORINTHIANS - Calleri28 dezembro 2024
SÃO PAULO, SP - 05.03.2022: SÃO PAULO FC X CORINTHIANS - Calleri28 dezembro 2024 -
 Character - WiKirby: it's a wiki, about Kirby!28 dezembro 2024
Character - WiKirby: it's a wiki, about Kirby!28 dezembro 2024 -
 Max Steel vs Blue Beetle by DBRanger09 on DeviantArt28 dezembro 2024
Max Steel vs Blue Beetle by DBRanger09 on DeviantArt28 dezembro 2024 -
Lolbit Swag28 dezembro 2024
-
 Cute stuff Cute little drawings, Cute icons, Cat drawing28 dezembro 2024
Cute stuff Cute little drawings, Cute icons, Cat drawing28 dezembro 2024 -
 Que palavras os russos usam para dizer 'dinheiro'? - Russia Beyond BR28 dezembro 2024
Que palavras os russos usam para dizer 'dinheiro'? - Russia Beyond BR28 dezembro 2024 -
 don't start now - dua lipa Song lyrics wallpaper, Don't start28 dezembro 2024
don't start now - dua lipa Song lyrics wallpaper, Don't start28 dezembro 2024
